Orthogonal Validation of NGS and Long-Read Sequencing



From Candidate to Confirmed: Orthogonal SV Validation with EGM
Electronic Genome Mapping (EGM) simplifies the detection of complex structural variants, overcoming challenges like repeats and duplications in both short- and long-read sequencing. Offering a fast, cost-effective, and genome-wide structural view, EGM helps teams quickly confirm variant calls, moving from "candidate" to "confirmed."
- Increase confidence: Orthogonal confirmation improves PPV and interpretation for both NGS and LRS pipelines.
- Resolve structure: Determine event class, size, orientation, and architecture (balanced vs. unbalanced; complex CNVs; repeat expansions).
- Reduce rework: Clarify structure first, then design one correct targeted assay (PCR or small targeted LRS) only if junction sequence is needed.
- Align with best practices: Supports industry expectations (e.g., ACMG) for orthogonal confirmation of potentially pathogenic calls.
- See what sequencing may miss: Detect or disambiguate large and complex events that remain unresolved after NGS/LRS.
EGM Can Add Clarity to the Following Cases
Why Choose EGM for Orthogonal Validation?
1. Accurate Detection and High Concordance with Long-Read Data
In the white paper, Electronic Genome Mapping for Confirmation of Long Structural Variants, we compare genome-wide SV calls exceeding 5 kb from EGM and PacBio HiFi in HG002. EGM data demonstrates a strong correlation to insertion and deletion calls made by PacBio HiFi. The data demonstrate EGM's precision in confirming PacBio's SV calls with nearly identical size estimates, underscoring its capability as an orthogonal validation method.
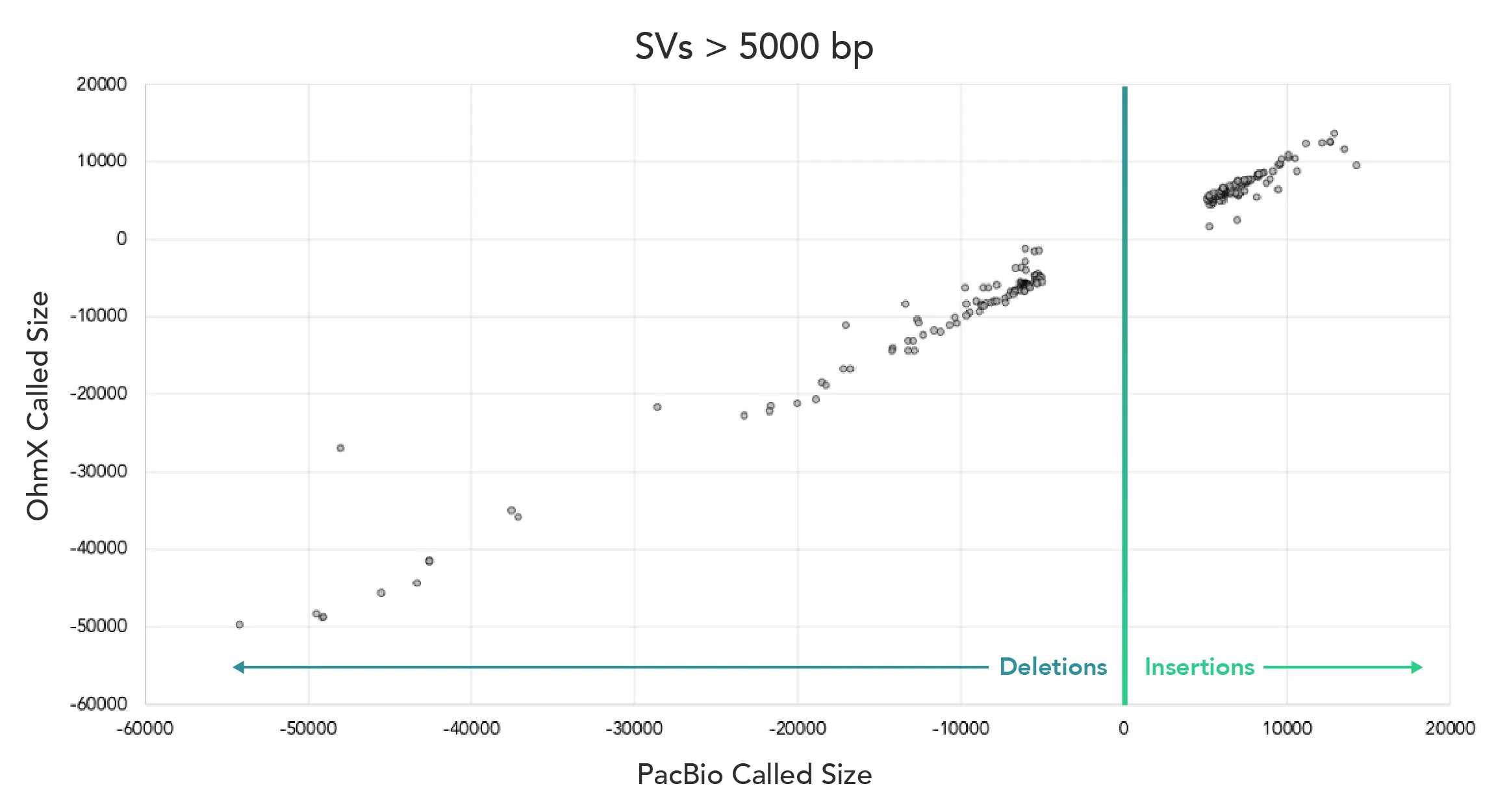
EGM validation of long-read SV calls visualised in Human Chromosome Explorer (HCE) software. Users can visually inspect genomic coordinates of interest and specific SV calls in HCE.
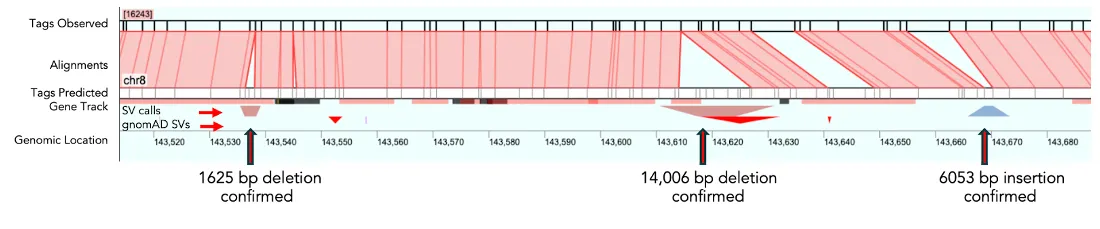
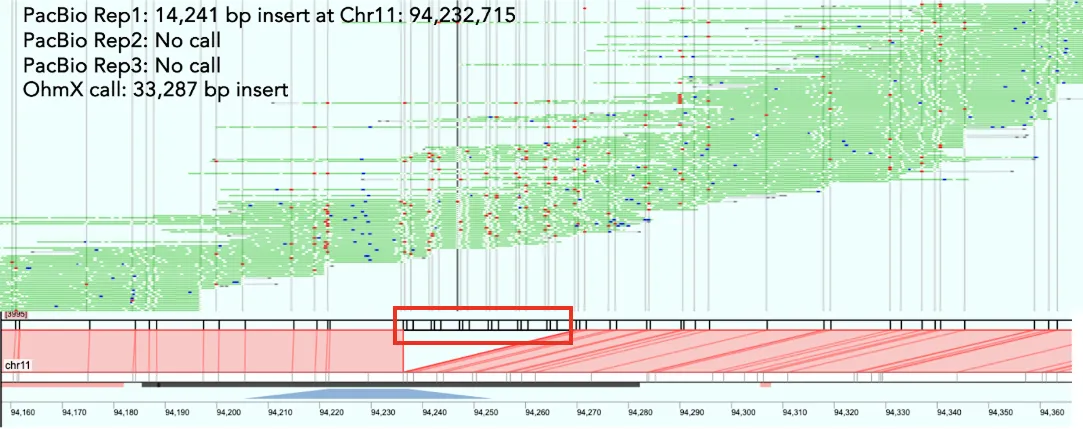
2. Clarification of Ambiguous SV Calls
Some SV calls may be ambiguous due to the complexity of genomic rearrangements, sequencing errors, or challenges in mapping repetitive and GC-rich regions. In these cases, EGM may provide clarity or additional context.
In one example below, EGM identified larger insertions, 33,287 bp, in an instance where only smaller or no SVs were called across PacBio replicates runs of HG002. In this case, EGM confirmed one of the PacBio calls but added additional information about the repetitive nature of the SVs. The pile up of EGM reads (in green) shows the high depth achieved across each region, which gives confidence in the call and provides the resolution of the repetitive motif in the red rectangles.
3. Low-Cost Validation Without Compromising Accuracy
Achieve robust results at a fraction of the cost compared to other orthogonal validation approaches, enabling scalable research and diagnostics.
Compared to PCR + Sanger Sequencing:
EGM eliminates the labor-intensive workflows and size limitations of PCR + Sanger, which are only practical for small SVs (<300–500 bp). While both methods offer low per-sample cost, EGM can validate SVs from 300 bp to megabases—offering far greater range and resolution in a similarly affordable, scalable format.
Compared to Additional Long-Read Sequencing:
Validating SVs by re-sequencing with a second long-read platform is costly and redundant. EGM provides orthogonal confirmation at 3–4x lower cost, while delivering high-resolution breakpoint accuracy without the need for deep re-sequencing or expensive computational analysis.
How It Works
Two cloud-based software pipelines are available on Google Cloud to confirm or clarify structural variants (SVs) that require orthogonal validation. To evaluate a candidate SV, users can conduct EGM analysis on the OhmX Platform, then upload the resulting data to one or both pipelines for further analysis.
The first pipeline, Human Chromosome Explorer (HCE), performs de novo genome assembly and genome-wide SV calling, delivering a comprehensive set of SV calls. Users can manually compare their candidate SV with HCE's output to confirm the SV or gain additional insights into the structure of the SV.
The second option is the SV Verify pipeline, which employs read-based mapping using a user-defined putative SV list. This pipeline specifically confirms or refutes the presence of the SV in the designated area.

Empower Your Research with Confidence
Accurate, reliable, and cost-efficient validation is just a step away. Discover how our cutting-edge technology can transform the way you approach SV analysis.
- Richards S, Aziz N, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24. doi: 10.1038/gim.2015.30.
Our Products
The state-of-the-art OhmX Platform uses electronic nano-detectors to deliver the highest resolution for whole genome structural variant analysis. You can now perform whole genome analysis of SVs down to 300bp in size—enabling insights into previously undetectable DNA variations.
Learn More

