Repeat Expansion Detection and Validation
.svg)


Why it Matters
Repeat expansions drive more than 60 neurological and neuromuscular disorders, yet they remain among the most difficult genomic features to study.1,2 PCR fails on large alleles, while short-read sequencing cannot map across long repetitive tracts because it relies on short molecules and inference.2,3 Even long-read platforms struggle to span GC-rich expansions at scale, and southern blotting continues to be slow, imprecise, and labor-intensive.3
Current Methods & Limitations
Existing tools require you to make compromises that leave critical genomic features like repeat expansions only partially resolved. This doesn't just slow down research — it creates gaps in our understanding and highlights the urgent need for new, complementary approaches to see the full picture.

How Nabsys Meets Your Needs
The OhmX Platform provides a robust and accessible solution for detecting and validating repeat expansions through its RepX Analysis for Repeat Expansion Disorders (RepX Analysis Pipeline)—overcoming many of the challenges that limit the ability of sequencing and legacy methods for analyzing repeat targets. Its unique capabilities deliver advantages in resolution, size detection, and reproducibility across diverse loci and cohorts.
- Resolution: EGM does not rely on amplification which enables detection of repetitive regions and motifs that are difficult for sequencing.
- Size: Ultra-long EGM reads enable resolution of normal, premutation, and full mutations and can span even the largest expansions.
- Reproducibility: End-to-end, the EGM workflow ensures reproducible analysis that can be applied across different loci and study cohorts.
Using EGM to Study Fragile X Syndrome
Fragile X syndrome, the most common inherited cause of intellectual disability, stems from large CGG repeat expansions in the FMR1 gene—regions that are difficult to resolve with sequencing alone. Electronic Genome Mapping (EGM) provides single-molecule resolution of these repeats, offering clearer insight into the genetic basis of the disorder and supporting improved diagnostics and therapeutic research.
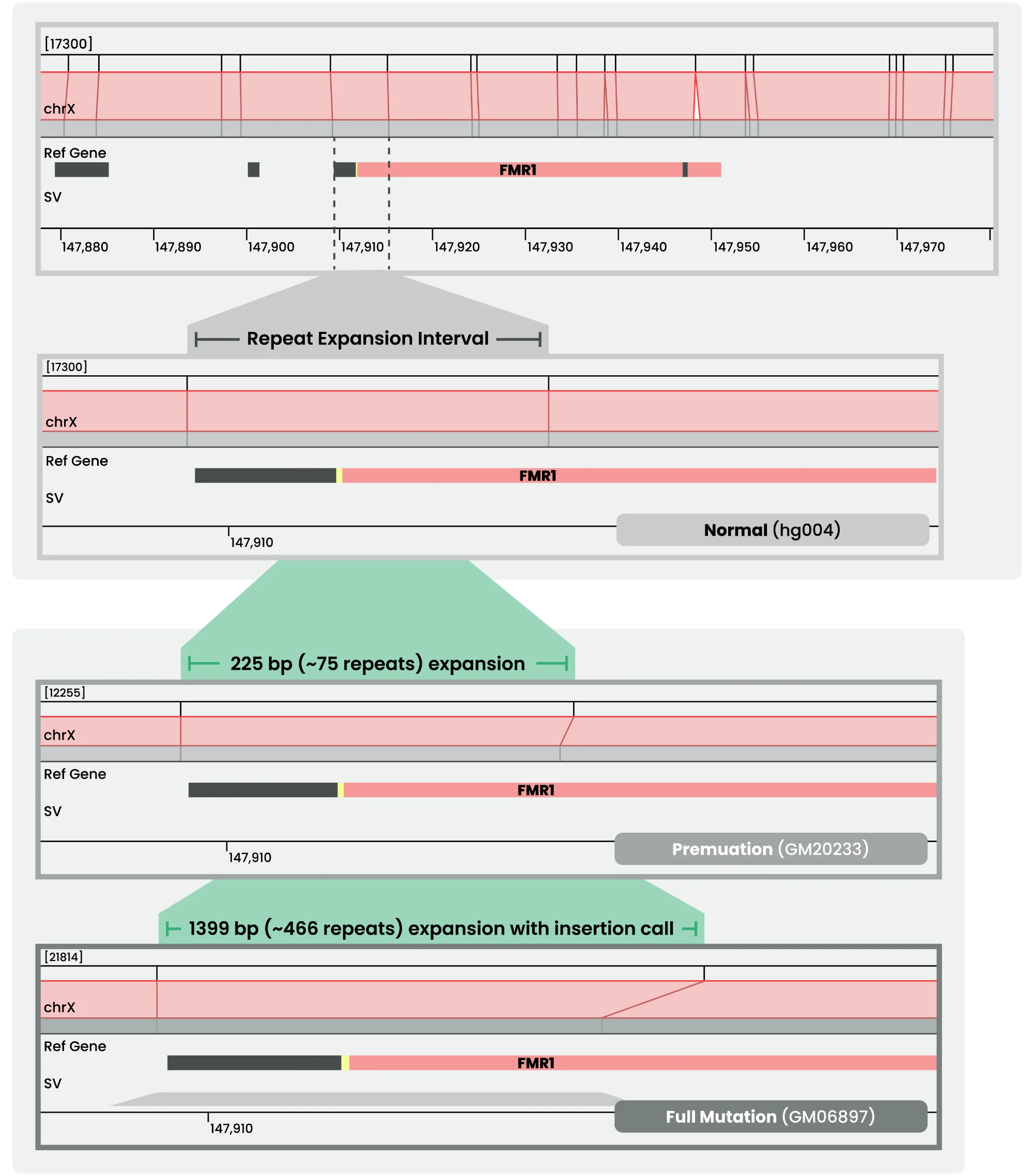
Resolving the FMR1 Repeat Expansion with EGM
For a detailed look at how the RepX Analysis Pipeline reveals CGG repeat expansions in FMR1, download our latest technical note. It walks through the workflow, data examples, and analysis approaches that demonstrate how EGM provides unmatched clarity in studying Fragile X and other repeat expansion disorders.

Download Fragile X Technical Note
Comprehensive Repeat Expansion Detection
The RepX Analysis Pipeline is a whole-genome assay that has the potential to screen for up to 30 repeats simultaneously in a single workflow, eliminating the need for multiple locus-specific assays. This comprehensive approach can streamline analysis, reduce turnaround time, and provide consistent resolution across diverse repeat types.
- Depienne,C., & Mandel, J.-L. (2021). 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? The American Journal of Human Genetics, 108(5), 764–785. https://doi.org/10.1016/j.ajhg.2021.03.011
- Stevanovski, I., Chintalaphani, S. R., Gamaarachchi, H., Ferguson, J. M., Pineda, S. S., Scriba, C. K., … Deveson, I. W. (2022). Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Science Advances, 8(9), eabm5386. https://doi.org/10.1126/sciadv.abm5386
- Miyatake,S., Koshimizu,E., Fujita, A., Doi, H., Okubo, M., Wada, T., … Matsumoto, N. (2022). Rapid and comprehensive diagnostic method for repeat expansion diseases using nanopore sequencing. NPJ Genomic Medicine, 7, 62. https://doi.org/10.1038/s41525-022-00331-y
Our Products
The state-of-the-art OhmX Platform uses electronic nano-detectors to deliver the highest resolution for whole genome structural variant analysis. You can now perform whole genome analysis of SVs down to 300bp in size—enabling insights into previously undetectable DNA variations.
Learn More

