Channeling Innovation: Advancements in Structural Variant Detection
Introduction
Genome mapping has expanded our understanding of biology, offering a powerful viewpoint for unraveling the structure and organization of DNA. While sequencing focuses on decoding the nucleotide sequence of DNA, mapping can reveal larger changes to genomic DNA.
Structural variants (SVs) are variants larger than 50 base pairs in size and affect a larger number of bases in the human genome than single-nucleotide variants (SNVs) and small indels, combined.1,2 Sequencing, particularly short-read sequencing (SRS), excels at detecting small variants like SNVs and indels, but comprehensive SV detection remains a challenge since many SVs are longer than SRS read lengths. However, these larger genomic variations are where genome mapping excels, offering clarity and context that sequencing alone can’t provide.
Enter Nabsys and its groundbreaking OhmX™ Platform—an innovative approach to genome mapping that redefines the possibilities. The platform uses a unique electronic nanochannel detection system, providing high precision and facilitating powerful insights into variation in the human genome.
In this post, we’ll explore the science behind Nabsys’ nanochannel technology, compare it to biological nanopore sequencing, and highlight the advantages that make it a game-changer in genomics.
How Nabsys Electronic Detection Works
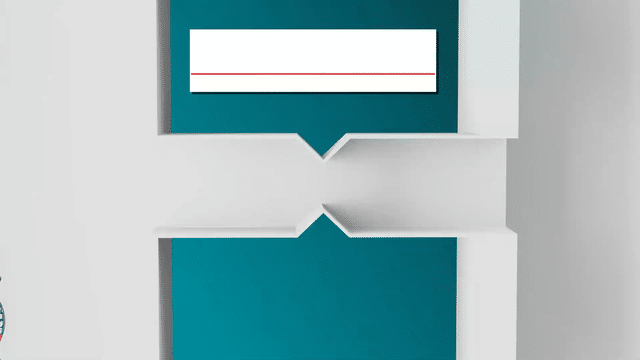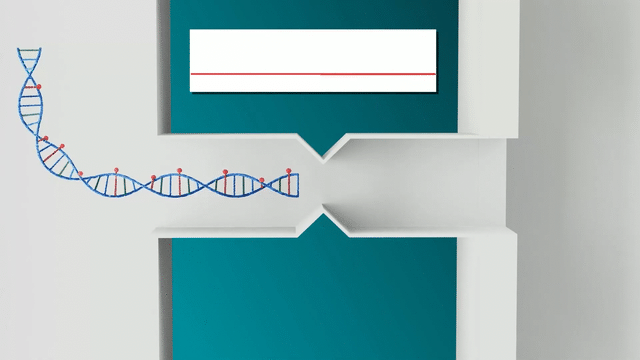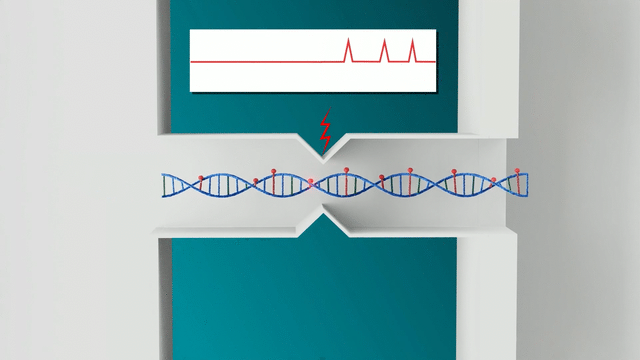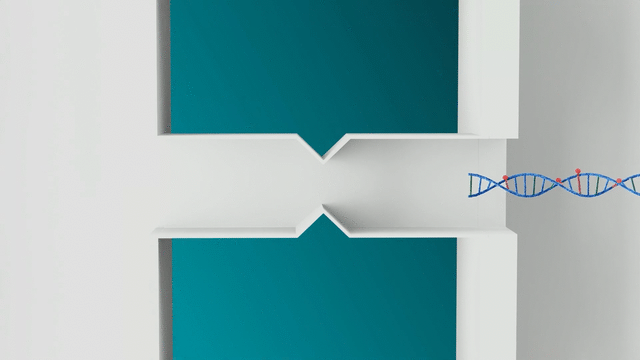
The Nabsys OhmX Platform relies on the electrophoresis of ultra-long DNA molecules through nanochannels. Electrodes are placed in the nanochannel and measure the voltage drop across the nanochannel as DNA molecules translocate through. When a molecule enters the channel, it partially displaces ions in solution, resulting in a measurable change in the voltage drop. This change indicates both the translocation of DNA and the positions of site-specific tags along each DNA molecule, allowing for accurate mapping to the human genome.
The OhmX Platform uses solid-state, silicon-based nanodetectors, which are stable at room temperature and offer the potential for reuse. For example, if you inject a low-quality sample into an OhmX Detector, you have the option to stop the run, flush the detector, and inject a different sample.
Additionally, EGM operates effectively at low DNA concentrations, minimizing DNA entanglement and maximizing yield, thus reducing sample preparation effort while maintaining long read lengths and high precision. This results in more comprehensive and accelerated data generation, offering researchers faster and more reliable insights.
To better understand the advantages of this approach, it’s helpful to compare it with one of today’s most widely used sequencing methods—nanopore sequencing.
How Nanopore Technology Works
Nanopore sequencing is one of the leading technologies in genomics, recognized for its ability to sequence DNA in real time. This method involves passing DNA molecules through tiny holes, or nanopores, embedded in a membrane. As the DNA moves through the nanopore, it disrupts the ionic current passing through, and the change in current identifies the sequence of the DNA.
One of the primary advantages of nanopore technology is its ability to provide real-time sequencing, enabling immediate analysis. This makes it particularly valuable for rapid applications like pathogen detection and clinical diagnostics. The portability of nanopore devices, ranging from benchtop to handheld models, also contributes to their popularity in laboratory and field settings.
Despite its advantages, nanopore sequencing is limited in its detection of large, complex SVs. The technology generally requires high DNA concentrations, which can cause the DNA molecules in the sample to become entangled. This entanglement negatively affects the sequencing yield, particularly with ultra-long DNA, resulting in incomplete data and increased costs to compensate. These limitations underscore the need for additional technologies, like electronic genome mapping (EGM), which provide a more precise and reliable solution for mapping complex genomic regions.
Benefits of Nanochannel Detection
The Nabsys OhmX Platform leverages the power of EGM through nanochannels, offering several key advantages over other methods like nanopore sequencing. These advantages make it particularly effective for mapping large genomic variations, including SVs, and pave the way for new insights in genomics.
1. Detection of Larger, More Complex SVs
Nanopore sequencing uses an electric field to drive DNA molecules through nanopores embedded in a membrane and relies on high DNA concentrations to maintain a high frequency of translocation through the nanopore. This concentration requirement increases the risk of DNA entanglement in the sample, which occurs when long strands of DNA intertwine with one another. These entangled DNA molecules can form a gel that separates them from the bulk solution and smaller strands of DNA, making it challenging to detect ultra-long DNA molecules.
In contrast, the Nabsys OhmX Platform applies an electric field across the entire fluidic system. This field guides DNA molecules into the nanochannels where detection occurs. The electric field facilitates DNA capture, enabling higher sensitivity, even at lower DNA concentrations. This lower concentration prevents DNA from becoming entangled, ensuring that the ultra-long DNA molecules that are critical for mapping SVs remain in solution and can be detected, thereby providing more reliable and precise mapping. This results in the ability to detect longer, more complex SVs, which is particularly important in cytogenetics and rare disease studies.
2. Detector Stability and Flexibility
A major drawback of nanopore technologies is their reliance on protein-based pores, which can be less stable than solid-state detectors and typically require refrigeration to maintain functionality. This dependence makes them costly and inconvenient to use, especially in settings with limited access to cold storage or during fieldwork scenarios.
EGM addresses these challenges with a solid-state detector that is far more stable and durable. While the OhmX workflow reagents require refrigeration, the detector does not, providing a more practical and robust solution. The solid-state detector is also highly resilient—even if a poor-quality or contaminated sample is introduced, the system can be easily flushed and cleaned, allowing the detector to be reused with a fresh aliquot of the same sample without any damage or significant downtime. This ease of maintenance makes the technology both efficient and cost-effective for a wide range of applications.
3. Speed and Efficiency
Nabsys’ innovative EGM technology delivers both cost-effectiveness and workflow efficiency. By leveraging scalable semiconductor fabrication and durable solid-state materials, the OhmX Platform reduces production costs compared to biologically derived nanopores. Its robust design also minimizes consumables usage and hands-on time, enabling full throughput in a single automated run. Together, these factors provide researchers with significant cost savings and faster, more iterative workflows.
Additionally, EGM streamlines some traditionally time-intensive nanopore workflows, cutting sample preparation and data collection times from days to overnight using advanced nanochannel systems.
For comparison, biological nanopore workflows typically require multiple reloads and manual wash steps to maximize yield, which increases hands-on time and extends run times to 60–72 hours. In contrast, the OhmX Platform reaches full throughput in a single load with a 16-hour run that is completely automated. This improvement not only conserves resources but also enables faster, iterative experimentation, addressing the evolving needs of genomics research.
Conclusion
The Nabsys OhmX Platform is redefining the possibilities of genome mapping with its innovative EGM technology. By leveraging control of an electric field and solid-state nanochannel detection, it overcomes many of the challenges faced by biological nanopore sequencing methods, particularly when it comes to detecting SVs.
With the ability to detect DNA at lower concentrations, avoid entanglement, and handle ultra-long DNA, the Nabsys OhmX Platform offers a level of precision and reliability that is crucial for understanding complex genomic regions. This approach not only enhances the accuracy of genome mapping but also opens the door for deeper insights into human health and disease.
Citations
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363-376. doi:10.1038/nrg2958
- Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464(7289):704-712. doi:10.1038/nature08516
.avif)

