Supercharging Repeat Expansion Detection with Electronic Genome Mapping
Repeat expansions are some of the most challenging regions of the genome to study, yet they play a central role in dozens of genetic disorders, including Huntington’s disease, Fragile X syndrome, some forms of amyotrophic lateral sclerosis (ALS), and spinocerebellar ataxias. Repeat expansions are stretches of DNA where a short sequence is repeated over and over, sometimes hundreds or thousands of times. The size of these repeats often determines whether someone is at risk, a carrier, or likely to develop the disease. Despite their clinical importance, methodical detection and characterization of repeat expansions remain significant hurdles in genomics research.
At Nabsys, we're helping researchers change that. Our electronic genome mapping (EGM) technology provides a unique and powerful method for detecting and sizing repeat expansions using the OhmX™ Platform.
Why These Repeats Matter
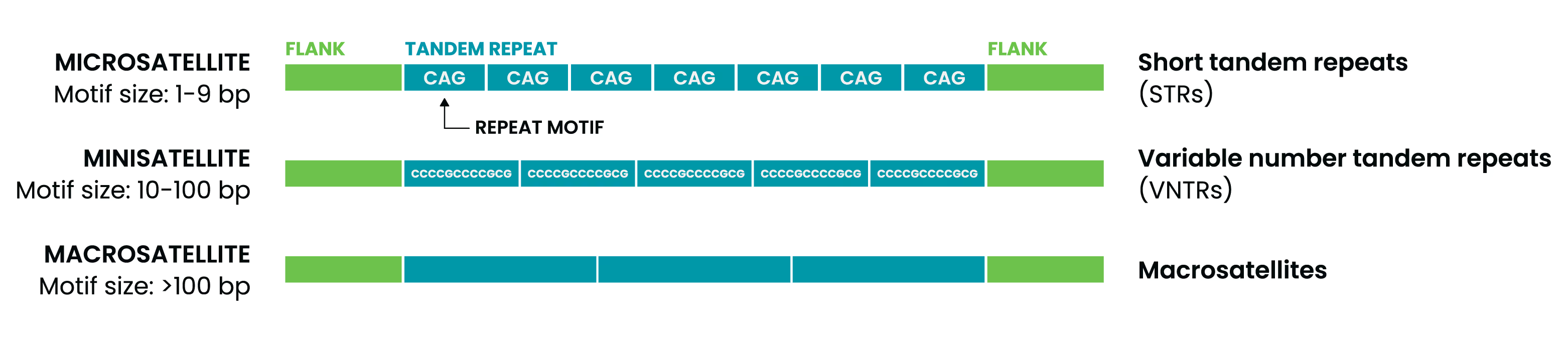
Repeat expansions are grouped into categories based on the length of the repeated unit:
- Microsatellites: 1–9 bp, also known as short tandem repeats (STRs)
- Minisatellites: 10–100 bp, includes variable number tandem repeats (VNTRs)
- Macrosatellites:> 100 bp
Many clinically relevant expansions fall under STRs or VNTRs. Their impact on human health is highly context-dependent. In many cases, expansions that reach or exceed pathogenic thresholds can:
- Inactivate neighboring genes through mechanisms such as promoter methylation
- Cause the production of toxic RNA species that interfere with normal cellular processes
- Lead to aberrant protein conformations, resulting in loss of function or aggregation
The links between specific repeat expansions and neurological or neuromuscular diseases underscore the need for methodologies that provide precise and reproducible measurements of repeat sizes and configurations.
What Makes Them So Hard to Detect?
Accurately resolving repeat expansions is a recognized technical challenge in genomics, primarily due to their size, instability, high GC content, and propensity for forming atypical secondary structures.
- Short-read sequencing is inherently limited in read length, making it unsuitable for spanning large expansions in a single read.
- Long-read sequencing has difficulty with specific motifs, such as GAA repeats, and may be constrained by throughput and cost factors.
- PCR-based assays often have allelic dropout of long expansions and are difficult to multiplex across many loci, limiting scalability and rish detection in heterogeneous samples.
- Southern blotting remains a widely-used technique for some expansions, but is labor-intensive and unsuitable for high-throughput and clinical applications.
These hurdles highlight the need for a technology capable of interrogating complex repetitive regions without sequence dropouts or amplification bias. In addition, labs that screen for repeat expansions using short-read NGS tools such as ExpansionHunter often need an orthogonal method to confirm their findings.
Unlocking Repeat Expansions with the OhmX Platform
The OhmX Platform, powered by EGM, directly detects ultra-long DNA molecules and provides an advanced alternative to current methodologies. This allows researchers to:
- Simultaneously interrogate multiple repeat expansions genome-wide
- Detect and size repeat expansions, including STRs and VNTRs, at the single-molecule level
- View both alleles and assess zygosity in diploid genomes
- Evaluate repeat expansions in the broader context of structural variants (SVs)—such as insertions, deletions, inversions, and translocations
EGM excels where other tools fall short—it doesn't require amplification or prior knowledge of the repeat locus, and it can span large repetitive regions that are otherwise inaccessible.
This capability has already been demonstrated at two challenging loci for repeat analysis. CGG expansions in FMR1 and GAA expansions in FXN have been successfully detected and sized on the OhmX Platform, with ultra-long reads spanning the full repeat regions and their flanking landmarks. In a single workflow, it is possible to distinguish premutation and full-mutation FMR1 alleles and to size difficult FXN expansions in intron 1, while also capturing nearby SVs that may influence locus behavior. These examples show how EGM overcomes the GC-rich and structurally complex nature of such regions—precisely the settings where sequencing-based assays typically falter.
Beyond FMR1 and FXN, EGM has the potential to be applied to additional STRs, VNTRs, and even macrosatellites where ultra-long single-molecule reads may provide new insight. A full and up-to-date list of candidate loci under evaluation is available on the Repeat Expansion Detection Application page.
Empowering Deeper Genomic Insights
Whether you're investigating classic STR disorders or looking at underexplored macrosatellite regions, the OhmX Platform, powered by EGM, provides the resolution and context needed to move your research forward—without the constraints for amplification bias or limited read lengths. Its integration of repeat expansion and SV analysis provides researchers and clinicians with an unprecedented toolkit for unraveling the role of these regions in human disease.
As the understanding of repeat expansions advances, novel insights and potential therapeutic targets will undoubtedly emerge. The OhmX Platform stands poised to support you in making these discoveries through precise, reliable, and efficient genomic analysis.
Discover what's possible when repeat expansions are no longer hidden.


